Motivation
We can parametrize Jenkins jobs with some standard parameter types (String, Text, Boolean, File etc.) as well as with parameters that are contributed from installed plugins (for example Active Choices).
All of these parameters are defined as Jenkins extension points, and we can use the Jenkins API to discover them.
From the Jenkins API documentation we learn that 'The actual meaning and the purpose of parameters are entirely up to users, so what the concrete parameter implementation is pluggable. Write subclasses in a plugin and put Extension on the descriptor to register them.'
How to find the Jenkins available parameter types
ep.each{
println it.getDisplayName()
}
Executing this script produces the following results on my Jenkins instance:
We essentially lookup in the Jenkins registered extensions, those that are of a specific class. Each parameter type has a corresponding ParameterDescriptor class that describes it. We then use the getDisplayName method to get the human readable names of these parameters (otherwise we get the class name of the plugin contributing the parameter) .
A look at a parametrized job structure
The ParametersDefinitionProperty list
ParametersDefinitionProperty, which in turns retains ParameterDefinitions, user would have to enter the values for the defined build parameters. Programmatic Job parametrization
| create a new job, lets call it 'MyTest01' | myJenkins=jenkins.model.Jenkins.instance myJenkins.createProject(FreeStyleProject,'MyTest01') job1=myJenkins.getJob("MyTest01") |
Add a ParametersDefinitionProperty to
the job
|
job1.addProperty(new
hudson.model.ParametersDefinitionProperty())
|
Now the job is
parametrized!
|
println
'IsParametized:'+job1.isParameterized()
|
Result
|
IsParametized:true
|
Now we have a parametrized job, but have not defined any parameters yet.
For simplicity, in this example we have used the null parameter constructor for ParametersDefinitionProperty. However, the constructor allows passing a list of parameter definitions, so if we add a list of ParameterDefinitions we can add the required project parameters!
Programmatic creation and parametrization of a free-style job
A complete example
We will now present a complete step-by-step example where we not only create a parametrized job but also add a couple of parameters.
To make this example even more interesting, one of the parameters will be an Active Choices parameter, and in the process we'll show how you can add the required Groovy script for the Active Choice parameter configuration.
The following code is a complete example of creating and parametrizing a free-style job programmatically.
Tasks
- Create a new Jenkins job
- Construct 2 parameters
- A simple String parameter
- An Active Choice parameter (with a secure groovy Script)
- Construct a ParametersDefinitionProperty with a list of 2 parameters
- Add the ParametersDefinitionProperty to the project
- Print some diagnostics
a
|
jenkins=jenkins.model.Jenkins.instance
jenkins.createProject(FreeStyleProject,'MyTest01')
job1=jenkins.model.Jenkins.instance.getJob("MyTest01")
|
b.i
|
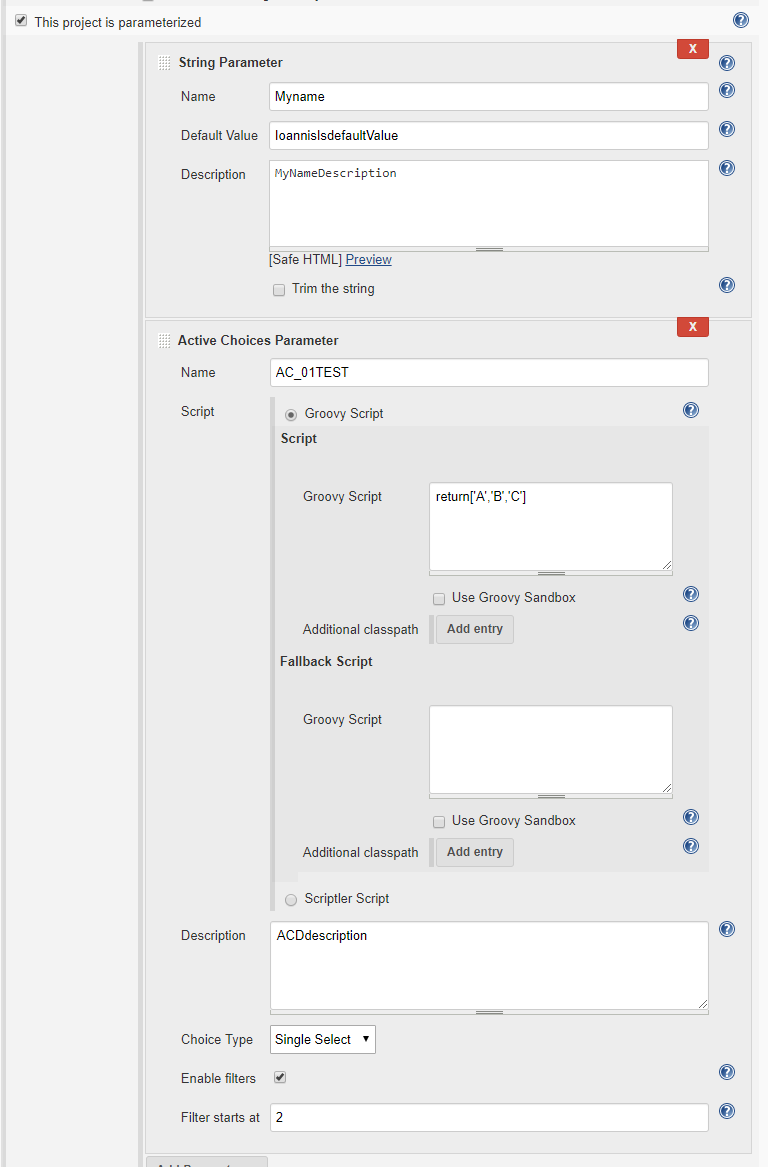
pdef1=new StringParameterDefinition('Myname',
'IoannisIsdefaultValue', 'MyNameDescription')
|
b.ii
|
sgs=new
org.jenkinsci.plugins.scriptsecurity.sandbox.groovy.SecureGroovyScript("""return['A','B','C']""",
false, null)
acScript=new
org.biouno.unochoice.model.GroovyScript(sgs,null)
pdef2=new org.biouno.unochoice.ChoiceParameter ('AC_01TEST',
'ACDdescription','boo123', acScript, 'PARAMETER_TYPE_SINGLE_SELECT', true,2)
|
c,d
|
job1.addProperty(new
hudson.model.ParametersDefinitionProperty([pdef1,pdef2]))
|
e
|
println 'IsParametized:'+job1.isParameterized()
println job1.properties
job1.properties.each{
println
it.value.class
println 'Descriptor:'+it.value.getDescriptor()
if(
it.value.class==hudson.model.ParametersDefinitionProperty){
println 'Job
Parameters'
println it.value.getParameterDefinitionNames()
it.value.getParameterDefinitionNames().each{pd->
println it.value.getParameterDefinition(pd).dump()
}
}
}
|
Result
|
IsParametized:true
[com.sonyericsson.hudson.plugins.metadata.model.MetadataJobProperty$MetaDataJobPropertyDescriptor@5ba29d96:com.sonyericsson.hudson.plugins.metadata.model.MetadataJobProperty@23d5994d,
com.sonyericsson.rebuild.RebuildSettings$DescriptorImpl@52e43430:com.sonyericsson.rebuild.RebuildSettings@56c0e80c,
hudson.model.ParametersDefinitionProperty$DescriptorImpl@1b2facb8:hudson.model.ParametersDefinitionProperty@6054a6c5]
class
com.sonyericsson.hudson.plugins.metadata.model.MetadataJobProperty
Descriptor:com.sonyericsson.hudson.plugins.metadata.model.MetadataJobProperty$MetaDataJobPropertyDescriptor@5ba29d96
class com.sonyericsson.rebuild.RebuildSettings
Descriptor:com.sonyericsson.rebuild.RebuildSettings$DescriptorImpl@52e43430
class hudson.model.ParametersDefinitionProperty
Descriptor:hudson.model.ParametersDefinitionProperty$DescriptorImpl@1b2facb8
Job Parameters
[Myname, AC_01TEST]
<hudson.model.StringParameterDefinition@125d74
defaultValue=IoannisIsdefaultValue trim=false name=Myname
description=MyNameDescription>
<org.biouno.unochoice.ChoiceParameter@35c83a1d
choiceType=PARAMETER_TYPE_SINGLE_SELECT filterable=true filterLength=2
visibleItemCount=1 script=GroovyScript [script=return['A','B','C'],
fallbackScript=] projectName=null randomName=boo123 name=AC_01TEST
description=ACDdescription>
|
Examine the configuration of the newly created project